Native chromatin immunoprecipitation (N-ChIP) of Schistosoma mansoni (5/3/2008)
© Christoph Grunau, Céline Cosseau, Abdelhalim Azzi, 2007,
This protocol is based on the protocol of David Umlauf. The authors are grateful to Jerome Buard for help reducing unspecific background.
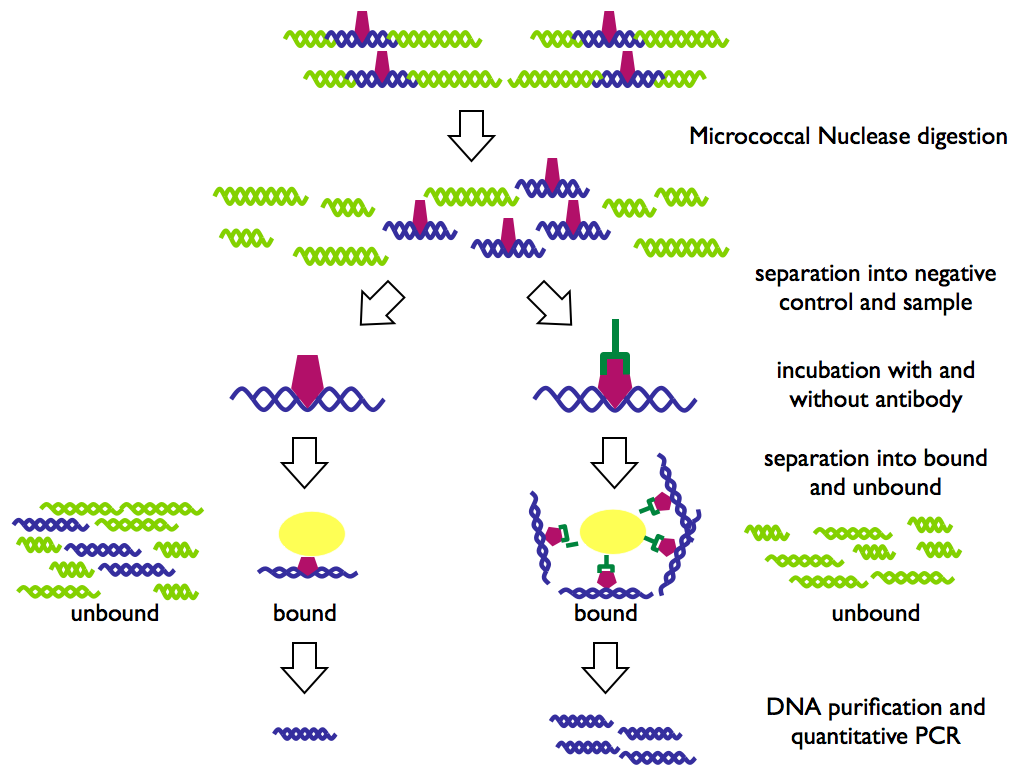
Before everything begins: prepare solutions...
- 1 M KCl, autoclave
- 5 M NaCl, filter and autoclave
- 1 M MgCl, autoclave
- 1 M Tris/Cl pH 7.4 - 7.6, autoclave
- 0.5 M EDTA, autoclave
- 1 M CaCl2, 10 ml, sterile filter or autoclave
- 100 mM DTT, 1ml (store at -20°C)
- Roche CompleteProtease Inhibitor (ref: 11 697 498 001)
- 2.5 M Sodium butyrate (Sigma B5887 1g) (store at 4°C)
- 25 mM PMSF in isopropanol, 10 ml (store at -20°C)
- 15 U/µl Micrococccal nuclease (MNase) (USB 70196Y) in sterile 50% glycerol, aliquot to ~10µl and store at -20°C
- Protein A - sepharose CL-4B (Sigma P3391 250mg) (store at 4°C)
- agarose gel loading buffer
- 20% SDS
- 20 g/l glycogen solution (store at -20°C)
- 2% NaN2 in water (store at 4°C)
- micro-dialysis units (Slide-a-Lyzer 3500 D cut-off, Pierce 69550). Note: You can also make your own (much cheaper) micro-dialysis units. For details see here (external link, not tested).
| preparation of protein A - sepharose |
250 mg Protein A - sepharose swells to approx. 1 ml gel and binds approx. 20 mg human IgG. You will need 50 µl of the protein A sepharose homogeneously mixed in its 5 ml water volume per ChIP. |
| optional if micro dialysis units are not available: preparation of dialysis tubing |
|
day 1...
- reserve centrifuge and cool down to 4°C
- prepare a 2% 0.5x TBE agarose gel with 20 µl slots
- preheat a water bath to exactly 37°C
- prepare the following solutions with autoclaved distilled water
| 2x base buffer | ||
| 6 ml | 1 M KCl | 60 mM final 1x |
| 0.3 ml | 5 M NaCl | 15 mM |
| 0.5 ml | 1 M MgCl2 | 5 mM |
| 20 µl | 500 mM EDTA | 0.1 mM |
| 1.5 ml | 1 M Tris/Cl | 15 mM |
| to 50 ml | water | |
| 2 | Roche protease inhibitor tabletts | |
| buffer1 (0.3 M sucrose) | |
| 2.58 g | sucrose |
| 12.5 ml | 2x base buffer |
| 50 µl | sodium butyrate |
| 100 µl | PMSF |
| 125 µl | DTT |
| to 25 ml | water |
| buffer 2 | |
| 10 ml | buffer 1 (0.3 M sucrose) |
| put on 37°C to allow NP40 to be pipetted into the buffer | |
| 80 µl | NP40 (cut pipette tip) |
| put on 37°C to fully dissolve NP40 and put on ice | |
| buffer 3 (1.2 M sucrose) for 3 cell samples | |
| 20.55 g | sucrose |
| 25 ml | 2x base buffer |
| 100 µl | sodium butyrate |
| 200 µl | PMSF |
| 250 µl | DTT |
| to 50 ml | water |
| MNase digestion buffer | |
| 1.1 g | sucrose |
| 0.5 ml | Tris/Cl |
| 80 µl | PMSF |
| 40 µl | MgCl2 |
| 20 µl | sodium butyrate |
| 10 µl | CaCl2 (essential for the enzyme) |
| to 10 ml | water |
| put at 37°C | |
| Dialysis buffer | |
| 1mM Tris/Cl, 200 µM EDTA, 200 µM PMSF, 5 mM sodium butyrate | |
| 50 µl | Tris/Cl |
| 20 µl | EDTA |
| 400 µl | PMSF |
| 100 µl | sodium butyrate |
| to 50 ml | water |
- put all buffer solutions on ice
(except MNase buffer)
- cell lysis
- aliquote 1500 sporocysts, miracidia or 10-20 adults (stored at -80°C or in liquid nitrogen)
- adults:
- remove excess liquid (if any) and resuspend in 1 ml buffer 1, add 1 ml buffer 2 (lysis buffer) and transfer to Dounce
- homogenize for 3 min with Dounce (pestle A) on ice
- put on ice 7 min
- sporocysts or miracidia:
- centrifuge into Eppendorf tubes, rinse storage tubes with PBS to recover all larvae
- centrifuge at 4000 rpm, 10 min, 4°C
- remove supernatant
- resuspend completely in 1 ml buffer 1
- do not add human lymphoblast cells as carrier
- add 1 ml buffer 2 (lysis buffer) and homogenize for 3 min with Dounce (pestle A) on ice
- put on ice 7 min
- fill 8 ml buffer 3 into a 50 ml corex centrifugation tube
- overlay the 8 ml buffer 3 with 1 ml cell suspension so that the tubes are ready for centrifugation 15 min (sporocysts) of 10 min (adults) after buffer 2 has been added to the cells
- disturb a little bit the interface
- use 2 corex tubes for the sporocysts sample that is in 2 ml buffer 1+2
- mark tubes at the exterior side (to know where to look for the nuclei)
- centrifuge 8500 rpm 20 min 4°C
- carefully remove supernatant
completely
- MNase digestion
- resuspend pellet in 1 ml MNase digestion buffer
- aliquot 500 µl of this suspension in 1.5 ml Eppendorf tubes
- add 1 µl MNase (15 U) and incubate 4 min at 37°C
- to stop the reaction add 20 µl 0.5 M EDTA to each 500 µl MNase digest and put the tube on ice
- centrifuge 13000 g 10 min 4°C
- transfer the supernatant to a new tube (S1) and keep the pellet (P1)
- store S1 at -20°C
- quantify chromatin in S1 by measuring OD at 260 nm in disposable cells against MNase buffer (In general we find about 50 µg/ml
DNA in the undiluted S1, OD260/280 values can be bad because there is a
lot of protein in the solution. DNA quantification is therefore not precise
but sufficient for reproducibility.)
- Dialysis of P1
- humidify Slide-a-Lyzer with 50µl dialysis buffer
- resuspend the pellet P1 in 100 µl dialysis buffer and dialyze overnight at 4°C against 50 ml dialysis buffer with gentle stirring
- the next day, transfer dialysed sample to Eppendorf tubes and...
- centrifuge 13000 g 10 min 4°C
- transfer the supernatant to a new tube and repeat the centrifugation 2 times
- supernatant is fraction S2
- yesterdays supernatant S1...
- in parallel with the dialyses sample, centrifuge 13000 g 10 min 4°C
- transfer the supernatant into a new tube and repeat this centrifugation 2 times
- these triple centrifugations are IMPORTANT! They reduce the unspecific background!
- use 50 µl of S1 and S2 for phenol/chlorofrom extraction, centrifuge and load 20 µl of supernatant on 2% 0.5x TBE gel (100V, 25 min)
- incubation with antibody
- Ideally, the antibody should be in excess over the protein you want to precipitate. The antigen/antibody ration must be determined experimentally for each antibody . Prepare a dilution series of your chromatin in MNase buffer starting with 20 - 40 µg DNA for histon ChIP.
- Add appropriate amounts of
stock solutions to generate the
antibody incubation buffer NaCl 150 mM Tris/Cl 20 mM sodium butyrate 20 mM EDTA 5 mM PMSF 100 µM - you can download an Excel worksheet for calculation here (v0.1) or here (v1.0)
- dissolve chromatin from S1 (and S2 if you have dialyzed) in 1 ml buffer
- add about 2 µg antibody
- incubate overnight at 4°C on a rotating wheel
day 2...
day 3...
- precipitation
- prepare 50 µl of protein A - sepharose for each tube
- wash the beads to remove NaN2: short spin, remove supernatant and replace with equal volume of sterile water
- add 50 µl of protein A - sepharose to each tube
- incubate at least 4 h at 4°C on a rotating wheel
- prepare washing buffers (10
ml / tube) and cool down to 4°C:
washing buffers A B C 50 ml 100 ml 200 ml 300 ml Tris/Cl 50 mM 2.5 ml 5 ml 10 ml 15 ml EDTA 10 mM 1 ml 2 ml 4 ml 6 ml sodium butyrate 5 mM 100 µl 200 µl 400 µl 600 µl washing buffer A NaCl 75 mM 750 µl 1.5 ml 3 ml 4.5 ml washing buffer B NaCl 125 mM 1.25 ml 2.5 ml 5 ml 7.5 ml washing buffer C NaCl 175 mM 1.75 ml 3.5 ml 7 ml 10.5 ml - centrifuge chromatin/antibody mixture 10 min 4°C 11600 g
- keep the supernatant in a 2 ml tube. This is the unbound fraction UB.
- resuspend the pellet in approx. 1 ml washing buffer A and transfer into a 15 ml Falcon tube containing 9 ml washing buffer A
- mix for 10 min on a rotating wheel at 4°C (speed 6)
- centrifuge 10 min 4000 rpm 4°C and pour off supernatant
- add 10 ml washing buffer B, mix for 10 min on a rotating wheel at 4°C and centrifuge 10 min 4000 rpm 4°C
- pour off supernatant
- add 10 ml washing buffer C, mix for 10 min on a rotating wheel at 4°C and centrifuge 10 min 4000 rpm 4°C
- pour off supernatant
- centrifuge 10 min 4000 rpm 4°C
- remove remaining supernatant completely
- resuspend pellet in 500 µl
elution buffer
elution buffer 10 ml 20 ml SDS (20% stock) 1 % 500 µl 1ml Tris/Cl 20 mM 200 µl 400 µl NaCl 50 mM 100 µl 200 µl EDTA 5 mM 100 µl 200 µl sodium butyrate 20 mM 80 µl 160µl PMSF 100 µM 40 µl 80 µl water to 10 ml to 20 ml - transfer suspension to a 1.5 ml Eppendorf tube
- incubate 15 min at RT on a rotating wheel
- centrifuge 10 min 11600 g 18°C
- transfer supernatant to a 1.5 ml Eppendorf tube
- This is the bound
fraction B.
- DNA extraction
- extract DNA with phenol/chloroform from fractions B and UB
- add 1 µl of a 20 g/l glycogen stock solution
- add NaCl to 250 mM (26 µl and 52 µl) and add 1 volume isopropanol
- put overnight at -20°C
- precipitate by centrifugation and wash with 70% ethanol
- dry the pellet and resuspend in 20 µl 10 mM Tris/Cl or qPCR grade water
- use 1 µl of this DNA for PCR in 25 µl reactions (quantitative real-time PCR) or 10 µl (PCR)
Ref.:
Cosseau C, Azzi A, Smith K, Freitag M, Mitta G, Grunau C. "Native chromatin immunoprecipitation (N-ChIP) and ChIP-Seq of Schistosoma mansoni: Critical experimental parameters." Mol Biochem Parasitol. 2009;166:70-6. [PubMed]
Umlauf D, Goto Y, Feil R. "Site-Specific Analysis of Histone Methylation and Acetylation" Methods Mol Biol. 2004;287:99-120. [PubMed]
O'Neill LP, Turner BM. "Immunoprecipitation of native chromatin: NChIP." Methods. 2003 Sep;31(1):76-82. [PubMed]